For GenBank & Database of the TYLCV isolates sequences Go to Links
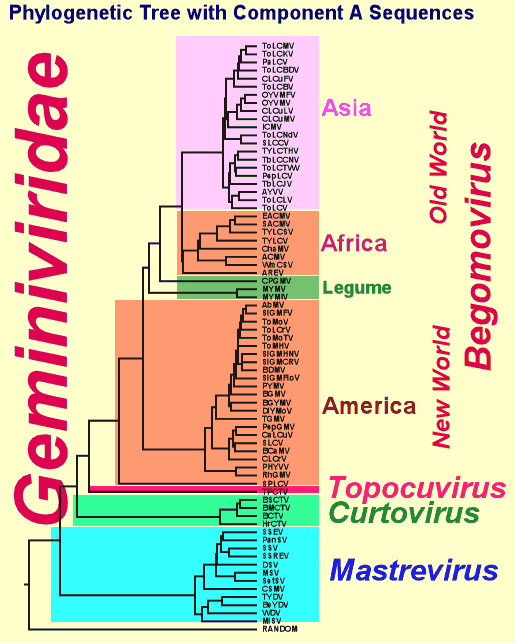
Taxonomy
Geminiviruses are plant viruses that belong to the family Geminiviridae,
first described by Goodman in 1977 (Goodman, 1977a, 1977b). Geminiviruses
are characterized by the unique Gemini shape of a fused icosahedral
viral particle.
The geminate virions consists a circular single-stranded DNA (ssDNA)
genome. The family Geminiviridae. is comprised of three genera, all
of which share similarities in
genome organization, insect transmission, and host range.
The genus Mastrevirus
Consists of geminiviruses with a monopartite genome, and the Mastreviruses
are transmitted by leafhoppers, in most cases by a single species in
a persistent, circulative, non-propagative manner.
The genus Curtovirus
Includes viruses with monopartite genomes, transmitted by leafhoppers
or treehoppers in a persistent, circulative, non-propagative manner.
Curtoviruses have very wide host ranges
The genus Begomovirus
Consists viruses with monopartite and bipartite genomes. Begomoviruses
are transmitted by whiteflies in a persistent, circulative, non-propagative
manner, and infect dicotyledonous plants. Bean golden mosaic virus (BGMV)
is the type species.
Geminivirus
Genome Organization
The geminivirus genome is organized in one (monopartite) or two (bipartite)
covalently closed, circular, ssDNA molecules of about 2.5 - 2.9 Kb (Lazarowitz,
1992).
The genes in monopartite and bipartite geminiviruses are arranged in
two divergent clusters 280 to 350 nucleotides each separated by the
intergenic region (IR)each. The single genomic component of monopartite
geminiviruses (mastreviruses and curtoviruses) contains all the information
necessary for virus replication and infectivity
(Lazarowitz, 1992; Hanley-Bowdoin et al., 1996). Bipartite begomoviruses
have seven genes distributed in the two genomic components designated
A and B. The A component contains genes involved in virus replication
and encapsidation, and the B component contains the genes involved in
virus movement (Lazarowitz, 1992). The A and B components each have
a common region, which consists of a block of approximately 200bp within
the IR (Sunter and Bisaro, 1991; Lazarowitz, 1992). The common regions
are virtually identical in sequence in a given bipartite begomovirus,
but are completely different in sequence among the other geminiviruses,
with the exception of a 30 nucleotide conserved region (stem loop) that
has been identified as the origin of replication (Sunter and Bisaro,
1991). The common region also contains two divergent promoters which
differentially regulate the temporal expression of the viral genes (Lazarowitz,
1992).
Begomoviruses
With the exception of tomato yellow leaf curl virus (TYLCV), which consists
of only one component (Lazarowitz, 1992), the genomes of begomoviruses
are made up of two components, each 2.5 - 2.8 Kb in size. The A component
of begomoviruses typically
has one gene in the virion sense and four genes in the complementary
sense (Lazarowitz,
1992).
AV1 (virion sense) encodes the coat protein, AC1 (complementary sense)
encodes the replication-associated protein (Rep) (Elmer et al., 1988),
and AC2encodes the transcriptional activator protein (TrAP) that transactivates
the expression of the coat protein gene and the BV1 movement gene of
the B component (Sunter and Bisaro, 1991). AC3 encodes the replication
enhancer protein (Ren) that regulates the virus replication rate, possibly
via the activation of an early gene (AV1) required for DNA synthesis
(Azzam et al., 1994). AC4 encodes a protein which is a determinant of
symptom expression in monopartite begomoviruses (Rigden et al., 1994).
The B component has two genes, designated BC1 and BV1. The product of
BV1 is localized in the cell nucleus and binds ssDNA, allowing the newly
formed virus genome to be transported to the cytoplasm (Pascal et. al
1993; Pascal et. al 1994). The BC1 product has been extracted from cell
wall and cellular membrane fractions, and its function is to increase
the exclusion limit of plasmodesmata to facilitate cell to cell movement
of the virus (Pascal et al., 1993). Both movement proteins define the
viral host range but only BC1etermines symptom severity and pathogenicity
in bipartite begomovirus (Ingham et al., 1995; Duan et al., 1997b).
Serological tests showed that all begomoviruses are related. In addition,
there is a
group of epitopes unique to the begomoviruses which infect crops in
the Old World
(Europe, Africa, and Australasia), and a distinct set of epitopes is
shared by
begomoviruses that infect crops in the New World (North, Central and
South America)
(Thomas et al., 1986).
Geminivirus
Replication
Geminivirus
replication strategy
The ssDNA genomes of geminiviruses replicate in the nucleus of infected
cells via a rolling circle mechanism using a dsDNA intermediate (Saunders
et al., 1991; Stenger et al., 1991). The replication process is analogous
to that used by ssDNA phages, such as ØX174 (Kornberg and Baker,
1992) and ssDNA plasmids such as pT181 and pC194 (Gros et al., 1987).
Moreover, sequence comparisons have shown that the geminiviral replication-associated
proteins are DNA binding proteins and are related to proteins involved
in the initiation of replication of some ssDNA plasmids (pMV158 family)
(Koonin and Ilyina, 1992).
The origin of replication includes a conserved 30 nucleotide putative
stem-loop element that is present in all geminiviruses (Revington et
al., 1989). A 5'- TAATATTAC- 3' motif, present in the stem-loop element,
is analogous to the A protein replication cleavage sequence in phage
1X140 (Saunders et al., 1991; Stenger et al., 1991; Arguello-Astorga
et al., 1994). The specific binding site for the replication associated-protein
in TGMV and SqLCV has been mapped to a region of about 60 nucleotides
upstream of the stem-loop element (Fontes et al., 1994; Lazarowitz et
al., 1992).
The Rep protein is a multifunctional protein that binds double-stranded
DNA, catalyzes cleavage and ligation of single-stranded DNA and forms
oligomers (Orozco and Hanley-Bowdoin, 1998). Rep protein initiates the
replication cycle by making a single stranded-cleavage of the virion
sense strand at the TAATATTAC sequence in the origin of replication.
After the DNA cleavage and strand transfer reaction at the origin of
replication, the Rep protein become covalently linked to the 5' end
of the cleaved DNA
(Laufs et al 1995). Luafs et al. (1995) demostrated that in TYLCV tyrosine-103,
located in the stem loop motif initiates DNA cleavage and is the physical
link between Rep protein and its origin DNA. Orozco and Hanley-Bowdoin
(1998) showed that the DNA binding motif in the Rep protein of TGMV
is located between amino acids 1 and 130.
The recognition of the origin of replication by the TGMV Rep protein
depends on a domain located between amino acids 121 and 200. The transcriptional
specificity is conferred primarily by amino acids 1 to 193 (Gladfelter
et al., 1997). The synthesis of ssDNA is regulated by the synergistic
activity of TrAp and Ren proteins that act as activator of transcription
and enhancer of replication, respectively (Sunter and Bisaro, 1991).
It has been proposed that geminiviruses depend on cellular factors to
complete their replicative cycles (Xie et al., 1999). A family of proteins
termed GRAB (for geminivius Rep-A binding) has been shown to bind to
the Rep A protein of wheat dwarf geminivirus. Two members of the family,
GRAB 1 and GRAB 2, have been characterized (Xie et al., 1999). The N-terminal
domain of GRAB proteins exhibit a significant amino acid homology to
the NAC domain present in proteins involved in plant development and
senescence.
Cytological
effects of geminivirus replication
The replication of geminiviruses induces micro-structural changes in
the nucleus of the host cells. Ultrastructural studies of Jatropha gossypifolia,
infected with the whitefly- transmitted begomovirus, jatropha mosaic
virus, showed fibrillar bodies and virus-like particles in the nuclei
of phloem-associated parenchyma cells and sieve elements (Kim et al.,
1986). The fibrillar bodies consists of two structural components with
different electron densities: the highly electron-dense beads and the
less electrondense matrix (Kim et al., 1986). Light microcopy studies
of leaf tissue of plants infected with BGMV and of lima bean golden
mosaic, euphorbia mosaic, malvaceous chlorosis, and rhyncosia mosaic
begomoviruses revealed nuclear inclusions that appear as large blue-
violet bodies when the tissue is stained with azure A (Christie et al.,
1986). These inclusions consist of aggregated virus particles. Small,
ring-shaped blue-violet, inclusions were also observed in the nuclei
of the phloem parenchyma cells. The nuclear inclusions were not observed
in stained tissues of non-infected host plants (Christie et al., 1986).
Pinner et al. (1990) observed four types of cytoplasmic inclusions induced
by maize streak virus and serologically related isolates: crystalline,
non-crystalline, sheet-like and open lattice. These distinctions enable
certain geminiviruses to be identified at the strain level.
Movement
The establishment of a virus infection depends upon the spread of the
virus through the plant host. The movement of the virus in the plant
occurs at two different levels: a) short distance cell- to -cell movement
and b) long-distance movement that involves delivery of the virus to
distal parts of the plant by the vascular system (Lazarowitz, 1992).
The BV1 and BC1 genes encode movement proteins in bipartite geminiviruses.
Studies of SqLCV (Pascal et al., 1994) and bean dwarf mosaic virus (Noueiry
et al., 1994) have shown that the BV1 and BR1 products act in a cooperative
manner to move the viral genome from the nucleus to the cytoplasm and
across the wall cell to a contiguous cell. It has been proposed that
BV1 is a nuclear shuttle protein. BV1 binds newly replicated ssDNA viral
genomes and transports them to the cytoplasm (Pascal et al., 1994: Sanderfoot
et al., 1996). Then, the BV1-genome complexes are directed to the cell
periphery through interactions with the BC1 product (Sanderfoot et al.,
1996; Sanderfoot and Lazarowitz, 1995). It has been suggested that the
BC1 protein allows the movement of BV1-genome complexes from one cell
to the next by increasing the exclusion limit of plasmodesmata (Sanderfoot
et al., 1996). The synthesis of BV1 is regulated at the transcriptional
level by AC2 transactivation (Sunter and Bisaro, 1991).
Whitefly-Transmission
of Geminiviruses
Bemisia tabaci B - biotype
Begomoviruses are transmitted by the sweetpotato whitefly, Bemisia tabaci
(Gennadius). B. tabaci was first described in the genus Aleyrodes in
1889 (Gennadius, 1889), and was first reported as a pest in 1919 in
India (Husain and Trehan, 1933). Since then, B. tabaci has been recognized
as a pest of crops in tropical and subtropical countries. B. tabaci
has a very wide host range, consisting of 500 species in 74 plant families
(Greathead, 1986). The whitefly is a vector of viruses in the Geminiviridae,
Potyviridae and Comoviridae families and the genera Carlavirus and Closterovirus.
Approximately 60 different geminiviruses have been reported to be transmitted
by B.
tabaci (Markam et al., 1994). In the New World before 1986, B. tabaci
was considered a pest of a limited number of crops (tobacco, cotton,
potato, bean, soybean), but by 1986 a sudden increase of the whitefly
population in ornamentals in Florida was observed (Osborne, 1988).
Shortly after that, whiteflies were reported in other crops in Florida
(Schuster et al., 1991), in California (Perring et al., 1991), and in
Texas, Arizona, Central America and South America (Brown, 1994). In
Florida the whitefly infestation was associated with silverleaf of squash
and irregular ripening of tomato (Maynard and Cantliffe, 1989). The
whitefly population causing these disorders was physiologically, behaviorally,
reproductively and genetically different from the population that was
present before 1989 in California and Arizona, and was first called
the “poinsettia strain”. Later, this whitefly became known
as the B strain or B biotype. Perring et al. (1991) suggested that A
and B strains were separate species, and thus, named the B strain (or
B biotype) the “silverleaf whitefly, B. argentifolii (Bellows
et al., 1994). The B biotype has a very wide host range, which has contributed
to the spread of geminiviruses to new hosts (Bedford et al., 1993) and
the outbreak of apparently new geminiviruses (Polston and Anderson,
1997).
Characteristics
of whitefly transmission
Whitefly-transmitted geminiviruses affect a wide variety of vegetable
crops worldwide.
In the 1930s, the first transmission of geminiviruses by whiteflies
was demonstrated with tobacco leaf curl virus and African cassava mosaic
virus in tobacco and cassava, respectively (Storey, 1934; Storey, 1936;
Storey and Nichols, 1938).
Geminivirus transmission by B. tabaci is circulative and non-propagative
(Duffus, 1987). Whiteflies can acquire and inoculate bipartite begomoviruses
in short periods of time (10 min), but the efficiency of acquisition
increases when the feeding period increases up to 24 h. Latent periods
of four to 21 h between virus acquisition and the ability of the whitefly
to transmit have been observed (Duffus, 1995) Studies of the transmission
of TYLCV, a monopartite begomovirus, showed that whitefly feeding periods
of 4 h or longer were necessary to achieve TYLCV transmission rates
near to 90% (Zeidan and Czeszcnek, 1991).
Hunter et al. (1998) established the location of tomato mottle begomovirus
(ToMoV) and cabbage leaf curl begomovirus (CaLCV) in various tissues
of B. tabaci Bbiotype by immunofluorescent labeling of viral coat protein
in freshly dissected whiteflies. Hunter et al. (1998) proposed the following
model for the movement of begomoviruses in the whitefly vector: virus
particles are ingested along with plant fluids into the whitefly esophagus
and foregut, after which nutrients and begomoviruses are concentrated
in the filter chamber. Begomovirus particles adsorb to specific sites
on the alimentary membrane or to sites along the anterior region of
the midgut. Begomovirus particles move out of these tissues into the
hemolymph, eventually invading the salivary glands.
Economic
Impact of Whitefly-Transmitted Geminiviruses
As early as the 1950s, there were reports of a correlation between the
presence of B. tabaci and plant diseases characterized by foliar malformation,
leaf curling, stunting and yellow mosaic in a variety of crops and weeds
in the Americas and the Caribbean basin (Brown and Bird, 1992). Many
of those diseases were later determined to be caused by geminiviruses
(Brown and Bird, 1992). Until the early 1990s, whitefly-transmitted
geminiviruses were primarily a problem in legume production in the Western
Hemisphere. Since then, high incidences of geminivirus diseases in tomato-producing
areas of Florida, the Caribbean, Mexico, Central America, Venezuela,
and Brazil have been reported (Polston and Anderson, 1997). Currently,
at least 17 geminiviruses have been reported infecting tomato in the
Americas and Caribbean region i. e. chino del tomato virus, tomato leaf
crumple virus, pepper huasteco virus, potato yellow mosaic virus, Sinaloa
tomato leaf curl virus, Texas pepper virus, pepper jalapeno virus, TYLCV,
ToMoV, serrano golden mosaic virus, tomato geminivirus BZ-Ub, tomato
geminivirus BZ-Ig, TGMV, tomato yellow mosaic virus, tomato yellow streak
virus, Tom GV1 virus, and Tom GV2 virus, with incidences ranging from
20 to 100% and causing crop losses up to 100% (Polston and Anderson,
1997).
Tomato geminiviruses have been reported to cause important losses in
the tomato producing areas of the Caribbean basin and Florida (Polston
and Anderson, 1997). The crop damage due to geminiviruses in the Dominican
Republic between 1988 and 1995 ranged from 5 to 95%, and the economic
losses from 1989 to 1995 were estimated at $50 million (Alvarez and
Abud-Antún, 1995). Tomato geminiviruses caused losses estimated
at $ 4.6 million in the Comayagua Valley of Honduras in 1992 (Caballero
and Rueda, 1993). In Venezuela, the area of tomato production was reduced
by 50% due to losses caused by tomato yellow mosaic virus (Salas and
Mendoza, 1995). In central America geminiviruses are thought to be responsible
for a significant portion of the crop losses estimated at $40 million
from 1989 to 1995 (Bird et al., 1995). The yields of the tomato crop
in Florida have been adversely affected by whitefly-transmitted geminiviruses.
In 1990 to 1991, crop losses due to ToMoV were estimated at $140 million.
(Schuster, 1992).In Florida and the Caribbean basin diseases caused
by whitefly-transmitted geminivirus diseases are also serious concerns
for many different crops such as beans, cassava, tobacco, potato, cotton,
pepper, squash, and cabbage (Polston and Anderson, 1997).
Readings
Azzam, O.,Frazer, J., De-La-Rosa, D., Beaver, J. S.,
Ahlquist, P., and Maxwell, D. P. 1994. Whitefly transmission and efficient
ssDNA accumulation of bean golden mosaic geminivirus require functional
coat protein. Virology 204: 289 - 296.
Arguello-Astorga,
G. R., Guevara-Gonzalez, R. G., Herrera-Estrella, L. R., and Rivera-
Bustamante, R. F. 1994. Geminivirus replication origins have a group
specific organization of iterative elements: A model for replication.
Virology 203: 90 - 100.
Alvarez,
P. A. and Abud-Antún, A. J. 1995. Reporte de Republica Dominicana.
CEIBA (Honduras) 36: 39 - 47.
Brown, J.
K. 1994. Current status of Bemisia tabaci as a plant and virus vector
in agroecosystems worldwide. FAO Plant Prot. Bull. 41: 3 - 32.
Brown, J.,
and Bird, J. 1992. Whitefly-transmitted geminiviruses and associated
disorders in the Americas and the Caribbean basin. Plant Dis. 76: 220
- 225.
Bird, J.,
Brown, J. K., Sosa, M., and Nazario, G. M. 1995.Reporte de Puerto Rico.
CEIBA (Honduras) 36: 37 - 38.
Caballero,
R., and Rueda, A. 1993. Las moscas blancas en Honduras. In: Las moscas
blancas (Homoptera: Aleyrodidae) en America Central y El Caribe. L.
Hilje and O. Arboleda (Eds.), CATIE, Turrialba, Costa Rica. pp 50 -
53.
Christie,
R. G., Ko, N. J., Falk, B. W., Hiebert, E., Lastra, R., Bird, J., and
Kim, K. S. 1986. Light microscopy of geminivirus-induced nuclear inclusion
bodies. Phytopathology 76: 124 - 126.
Duan, Y.
P., Powell, C. A., Purcifull, D. E., Broglio, P., and Hiebert, E. 1997a.
Phenotypic variation in transgenic plants expressing mutated geminivirus
movement/pathogenicity (BC1) proteins. Mol. Plant-Microbe Interact.
10: 1065 - 1074.
Duan, Y.
P., Powell, C. A., Webb, S. E., Purcifull, D. E., and Hiebert, E. 1997b.
Geminivirus resistance in transgenic tobacco expressing mutated BC1
protein. Mol. Plant-Microbe Interact. 10: 617-623.
Duffus,
J. E. 1987. Whitefly transmission in plant viruses. In Current Topics
in Vector of squash leaf curl virus (SqLCV) Research, Vol. 4 K. F. Harris
(Ed.), Springer Verlag, New York, pp. 73 - 91.
Duffus,
J. E. 1995. Whitefly-borne viruses. In Bemisia: 1995 Taxonomy, Biology,
Damage, Control and Management, D. Gerling, R. T. Mayer (Eds.), Intercept
Ltd., U K, pp. 257 - 258.
Elmer, J. E., Brand, L., Sunter, G.,Gardiner, W. E., Bisaro, D. M.,
and Rogers, S. G. 1988. Genetic analysis of the tomato mosaic golden
virus II. The product of the AL1 coding sequence is required for replication.
Nucleic Acids Res. 16: 7043 - 7060.
Fontes,
E. P. B., Gladfelter, H. J., Schaffer, R. L., Petty, I. T. D., and Hanley-Bowdoin,
L.1994. Geminivirus replication origin have a modular organization.
Plant Cell 6: 405 - 416.
Gros, M.
F., Te Riele, H., and Erlich, S. D. 1987. Rolling replication of the
singlestranded plasmid pC194. EMBO J. 6: 3863 - 3869.
Goodman,
R. M. 1977a. Single stranded-DNA genome in a whitefly-transmitted plant
virus. Virology 83: 171 - 179.
Goodman,
R. M. 1977b. A new kind of virus is discovered. Illinois Research 19:
5.
Greathead,
A. H. 1986. Host plants. In Bemisia tabaci-A literature Survey. M. J.
W. Cock (Ed.), CAB International Institute of Biological Control, Silwood
Park, Ascot, Berks., UK, pp. 17 - 25.
Gennadius,
P. 1889. Disease of tobacco plantations in the Trikonia. The aleurodid
of tobacco. Ellenike Georgia 5: 1 - 3 (in Greek).
Gladfelter,
H. L., Eagle, P. A., Fontes, E. P., Batts, L., and Hanley-Bowdoin, L.
1997. Two domains of AL1 protein mediate geminivirus origin recognition.
Virology 8: 186 - 197.
Hunter,
W. B., Hiebert, E., Webb, S. E., Tsai, J. H., and Polston, J. E. 1998.
Location of geminiviruses in the whitefly Bemisia tabaci (Homoptera:
Aleyrodide). Plant Dis. 82: 1147 - 1151.
Husain,
M.A., and Trehan, K. N. 1993. Observations on the life-history, bionomics
and control of white-fly of cotton (Bemisia gossypiperda M.& L.).
Indian Journal of Agricultural Science 3: 701 - 753.
Hanley-Bowdoin,
L., Eagle, P. A., and Orozco, B. M. 1996. Geminivirus replication. In:
Stacey G, Mullin B, Gesshoff P M (Eds) Biology of plant-microbe interactions.
Int. Soc. Mol. Plant-Microbe Interactions, St. Paul, MN, pp 287 - 292.
Ingham,
D. J., Pascal, E., and Lazarowitz, S. G. 1995. Both geminivirus movement
proteins define viral host range, but only BL1 determines viral pathogenicity.
Virology 207: 191 - 204.
Kornberg,
A., and Baker, T. A. 1992. DNA replication. 2nd ed. Freeman, New York.
Koonin,
E. V., and Ilyina, T. V. 1992. Geminivirus replication proteins are
related to prokaryotic plasmid rolling circle DNA initiation proteins.
J. Gen. Virol. 73: 2763 - 2766.
Kim, K.
S., Bird, J., Rodriguez, R. L., Martin, E. M., and Escudero, J. 1986.
Ultrastructural studies of Jatropha gossypifolia infected with jatropha
mosaic virus, a whitefly-transmitted geminivirus. Phytopathology 76:80
- 85.
Lazarowitz,
S. G. 1992. Geminiviruses: Genome structure and gene function. Crictic
Rev. Plant Sci. 11: 327 - 349.
Luafs, J.,
Schumacher, S., Geisler, N., Jupin, I., and Gronenborn, B. 1995. Identification
of the nicking tyrosine of geminivirus Rep protein. FEBS letter 18:
258 - 2262.
Markam,
P. G., Bedford, S. L., and Pinner, M. S. 1994. The transmission of geminivirus
by Bemisia tabaci. Pestic. Sci. 1994, 42: 123 - 128.
Maynard,
D. N., and Cantliffe, D. J. 1989. Squash silverleaf and tomato irregular
ripening: new vegetable disorders in Florida. University of Florida,
IFAS, Florida Cooperative Extension Service, Vegetable Crops Fact Sheet
VC-37.
Noueiry,
A. O., Lucas, W. J., and Gilbertson, R. L.1994. Two proteins of a plant
DNA virus coordinate nuclear and plasmodesmatal transport. Cell 76:
925 - 932.
Orozco,
B. M., and Hanley-Bowdoin L. 1998. Conserved sequence and structural
motifs contribute to the DNA binding and cleavage activities of a geminivirus
replication protein. J. Biol. Chem. 18: 24448 - 24456.
Osborne,
L. S. 1988. The not so sweet sweetpotato whitefly. Florida Foliage.
May, 8 - 15.
Pascal,
E., Goodlove, P. E., Wu, L. C., and Lazarowitz, S. G. 1993. Transgenic
tobacco plants expressing the geminivirus BL1 protein exhibit symptoms
of viral disease. Plant Cell 5: 795 - 807.
Pascal,
E., Sanderfoot, A. A., Ward, B. M., Medville, R., Turgeon, R., and Lazarowitz,
S. G. 1994. The geminivirus BR1 movement protein binds single stranded
DNA and localizes to the cell nucleus. Plant Cell 6: 995 - 1006.
Perring,
T. M., Cooper, A. D., Rodriguez, R. J., Farrar, C. A., and Bellows,
T. S. 1991. Identification of a whitefly species by genomic and behavioral
studies. Science 259: 74 - 77.
Polston,
J. E., and Anderson P. K. 1997. The emergence of whitefly-transmitted
geminiviruses in tomato in the Western Hemisphere. Plant Dis. 81: 1358
- 1369.
Polston,
J. E., Chellemi, D.O., Schuster, D. J., McGovern, R. J., and Stanley,
P. A. 1996. Spatial and temporal dynamics of tomato mottle virus and
Bemisia tabaci (Genn.) in Florida tomato fields. Plant Dis. 80:1022-1028.
Polston,
J. E., Hiebert, E., McGovern, R. J., Stansly, P. A., and Schuster, D.
J. 1993. Host range of tomato mottle virus, a new geminivirus affecting
tomato in Florida. Plant Dis. 77: 1181 - 1184.
Revington,
G. N., Sunter, G., and Bisaro, D. M. 1989. DNA sequences essential for
replication of the B genome component of tomato golden mosaic. Plant
Cell 1: 985 -992.
Rigden,
J. E., Krake, L. R., Rezaian, M. A., and Dry, I. B. 1994. ORF C4 of
tomato leaf curl virus is a determinant of symptom severity. Virology
204: 847 - 850.
Sunter,
G., and Bisaro, D. M. 1991. Transactivation in a geminivirus: AL2 gene
product is needed for coat protein expression. Virology 180: 416 - 419.
Saunders,
K., Lucy, A., and Stanley, J. 1991. DNA forms of the geminivirus African
cassava mosaic virus consistent with a rolling circle mechanism of replication.
Nucleic Acids Res. 19: 2325 - 2330.
Stenger,
D. C., Revington, G. N., Stevenson, M. C., and Bisaro, D. M. 1991. Replicational
release of geminivirus genomes from tandemly repeated copies: evidence
for rolling-circle replication of a plant vial DNA. Proc. Natl. Acad..
Sci. USA 88: 8029 - 8033.
Salas, J.,
and Mendoza, O. 1995. Reporte de Venezuela. CEIBA 36: 49 - 50.
Sanderfoot,
A. A., Ingham, D. L. and Lazarowitz, S. G. 1996. A viral movement protein
as a nuclear shuttle. The geminivirus BR1 movement protein contains
domains essential for interaction with BL1 and nuclear localization.
Plant Physiol. 110: 23- 33.
Sanderfoot,
A. A., and Lazarowitz, S. G. 1995. Cooperation in viral movement: The
geminivirus BL1 movement protein interacts with BR1 and redirects it
from the nucleus to the cell periphery. Plant Cell 7: 1185 - 1194.
Saunders,
K., Lucy, A., and Stanley, J. 1991. DNA forms of the geminivirus African
cassava mosaic virus consistent with a rolling circle mechanism of replication.
Nucleic Acids Res. 19: 2325 - 2330.
Sunter,
G., and Bisaro, D. M. 1991. Transactivation in a geminivirus: AL2 gene
product is needed for coat protein expression. Virology 180: 416 - 419.
Storey,
H. H., and Nichols, R. F. W. 1938. Studies on the mosaic of cassava.
Ann. Appl. Biol. 25: 790 - 806.
Storey,
H. H. 1934. Report of the plant pathologist. East Afr. Agric. Res. Stn.
Amani Annu. Rep. 6: 1 - 10.
Storey,
H. H. 1936. Virus diseases of East African plants. VI. A progress report
on studies of diseases of cassava. East Afr. Agric. J. 2: 34 - 39.
Thomas,
J. E., Massalski, P. R., and Harrison, B. D. 1986. Production of monoclonal
antibodies to African cassava mosaic virus and differences in their
reactivities with other whitefly-transmitted geminiviruses. J. Gen.
Virol. 67: 2739 - 2748.
Xie, Q.,
Sanz-Burgos, A. P., Guo, H., Garcia, J. A. and Gutierrez, C. 1999. GRAB
proteins, novel members of the NAC domain family, isolated by their
interaction with geminivirus protein. Plant Mol. Biol. 39: 647 - 656.
Zeidan,
M.,and Czescnek, H. 1991. Acquisition of tomato yellow leaf curl virus
by the whitefly Bemisia tabaci. J. Gen. Virol. 72: 2607 - 2614.
Prepared
by Mohd. K. Abhary (mohammedjabri@hotmail.com)