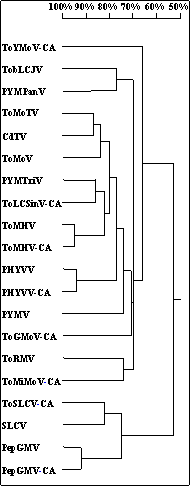
Fig. 1. Cladogram showing the relationships among 20 begomoviruses, based
on 805 nucleotides of the N-terminus of the Rep gene. Sequences were analyzed
by the Observed Divergency Distance Method of the Phylogenetic Analysis
Homology Tree module of DNAMAN software. Vertical distances are arbitrary,
and horizontal distances are in proportion to the number of nucleotide
differences between branch nodes. The % nucleotide differences are presented
by the scale at the top of the figure. The eight identified begomoviruses
in Central America: PepGMV, PHYVV, ToLCSinV, ToMHV, ToMiMoV, ToGMoV, ToSLCV,
and ToYMoV, were compared with other known begomoviruses: Chino del tomate
virus (CdTV, NC_003830), Pepper golden mosaic virus (PepGMV, NC_004101),
Pepper huasteco virus (PHYVV, NC_001359), Potato yellow mosaic Panama
virus (PYMPanV, Y15034), Potato yellow mosaic Trinidad virus (PYMTriV,
AF039031), Potato yellow mosaic virus (PYMV, NC_001934), Squash leaf curl
virus (SLCV, M38183), Tobacco leaf curl Japan virus (TbLCJV, AB079689),
Tomato mosaic Havana virus (ToMHV, NC_003867), Tomato mottle Taino virus
(ToMoTV, AF012300), Tomato mottle virus (ToMoV, NC_001938), Tomato rugose
mosaic virus (ToRMV, NC_002555). The acronyms of the eight viruses detected
in this study were labeled at the end with CA for Central America.